bioinfo-statistics
25.02.07 몇 가지 Drug prediction 논문 overview 본문
1. Drug-target interaction 관련 논문들
https://www.nature.com/articles/s41467-017-00680-8


drug, protein의 다양한 feature 를 input data에 포함. drug-protein interaction을 예측함. 단, 2017년 논문이어서 더 최신 논문이 있는지 알아볼 것
https://www.nature.com/articles/s41467-024-46089-y

Many machine learning applications in bioinformatics currently rely on matching gene identities when analyzing input gene signatures and fail to take advantage of preexisting knowledge about gene functions. To further enable comparative analysis of OMICS datasets, including target deconvolution and mechanism of action studies, we develop an approach that represents gene signatures projected onto their biological functions, instead of their identities, similar to how the word2vec technique works in natural language processing. We develop the Functional Representation of Gene Signatures (FRoGS) approach by training a deep learning model and demonstrate that its application to the Broad Institute’s L1000 datasets results in more effective compound-target predictions than models based on gene identities alone. By integrating additional pharmacological activity data sources, FRoGS significantly increases the number of high-quality compound-target predictions relative to existing approaches, many of which are supported by in silico and/or experimental evidence. These results underscore the general utility of FRoGS in machine learning-based bioinformatics applications. Prediction networks pre-equipped with the knowledge of gene functions may help uncover new relationships among gene signatures acquired by large-scale OMICs studies on compounds, cell types, disease models, and patient cohorts.
compoud-target prediction에 대한 논문. 특히 gene identity가 아닌 gene signiture (gene function)을 projection했다는 것이 차별점.
https://www.nature.com/articles/s41467-024-54440-6


protein-ligand docking prediction 에 관한 논문. 특히 complex interaction modeling에 집중한 것이 차별점
최근에 protein structure prediction을 넘어 protein-protein interaction prediction에 관심이 높아지고 있는 것 같던데 그것과 관련있는 것인가?
2. Drug response (side-effect, mode-of-action) 관련 논문들
https://www.nature.com/articles/nature11159


Side-effect (Drug가 다른 protein target에 영향을 주는 것) prediction. Nature에 나왔으나 2012년 논문임. 더 최신 논문이 있는지 찾아보기.
https://www.nature.com/articles/s42256-021-00408-w


(문제)
Data discrepancy between preclinical and clinical datasets poses a major challenge for accurate drug response prediction based on gene expression data. Different methods of transfer learning have been proposed to address such data discrepancy in drug response prediction for different cancers. These methods generally use cell lines as source domains, and patients, patient-derived xenografts or other cell lines as target domains; however, it is assumed that the methods have access to the target domain during training or fine-tuning, and they can only take labelled source domains as input. The former is a strong assumption that is not satisfied during deployment of these models in the clinic, whereas the latter means these methods rely on labelled source domains that are of limited size.
(해결)
To avoid these assumptions, we formulate drug response prediction in cancer as an out-of-distribution generalization problem, which does not assume that the target domain is accessible during training. Moreover, to exploit unlabelled source domain data—which tends to be much more plentiful than labelled data—we adopt a semi-supervised approach. We propose Velodrome, a semi-supervised method of out-of-distribution generalization that takes labelled and unlabelled data from different resources as input and makes generalizable predictions.
(해결 방법 설명)
Velodrome achieves this goal by introducing an objective function that combines a supervised loss for accurate prediction, an alignment loss for generalization and a consistency loss to incorporate unlabelled samples. Our experimental results demonstrate that Velodrome outperforms state-of-the-art pharmacogenomics and transfer learning baselines on cell lines, patient-derived xenografts and patients. Finally, we showed that Velodrome models generalize to different tissue types that were well-represented, under-represented or completely absent in the training data. Overall, our results suggest that Velodrome may guide precision oncology more accurately.
Organoid나 mouse model에서의 임상 결과로 patient 결과 예측 하는 것에 관심이 있어서 흥미로움. 특히 training에 필요한 patient 결과가 없을 가능성이 높다는 것이 가장 큰 문제 중 하나일텐데, 그것을 해결했다는 것에서 관심이 감.
논문 내용에 대한 챗지피티 링크: https://chatgpt.com/share/67f19f18-36dc-8004-a732-4c4e832138f1
https://www.nature.com/articles/s41587-024-02524-5

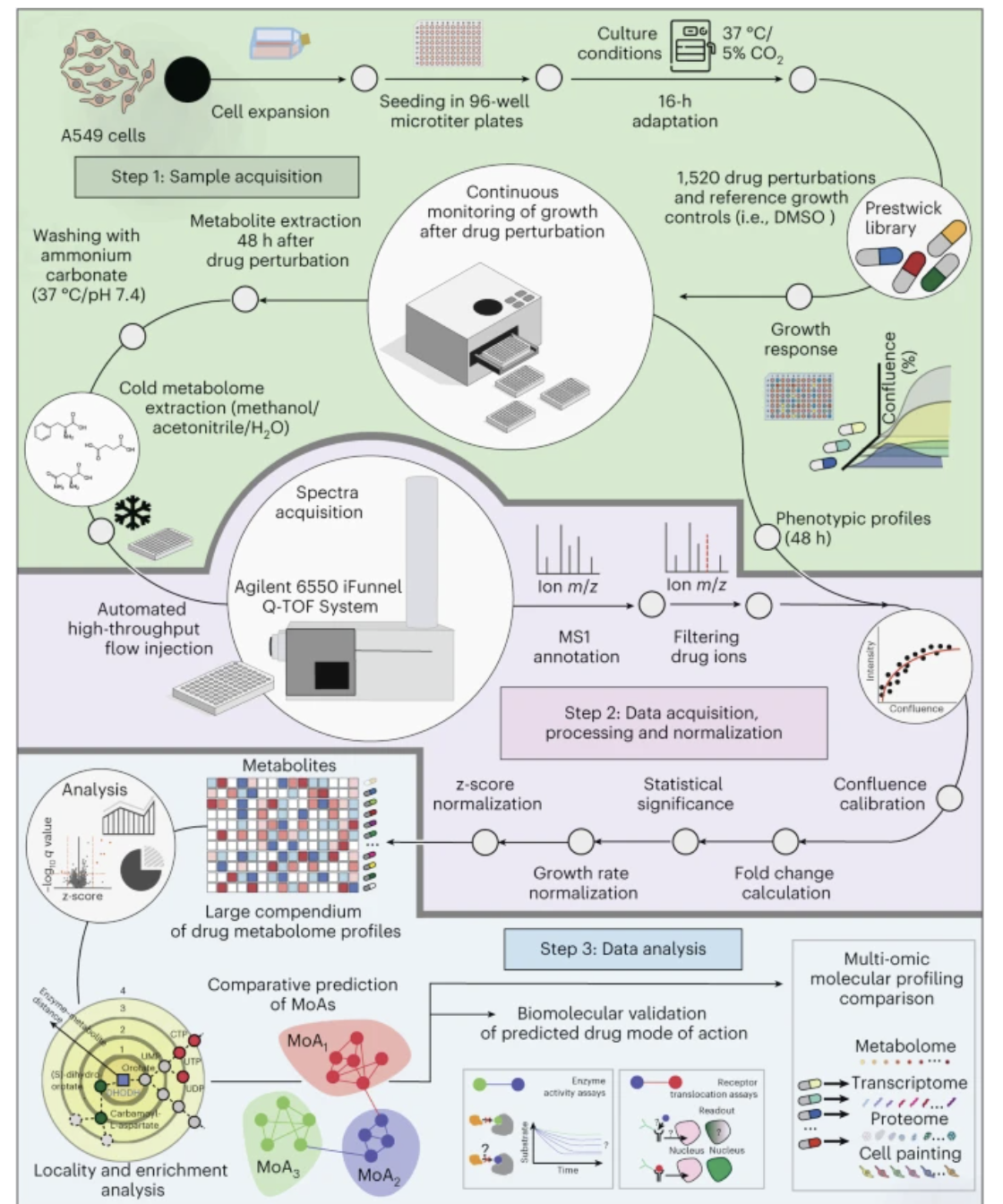
drug 처리 -> metabolite screening 데이터를 가지고 drug의 mode of action을 예측함.
https://www.nature.com/articles/s41587-022-01539-0


drug 처리 -> proteome screening 데이터를 가지고 drug의 mode of action을 예측함.



